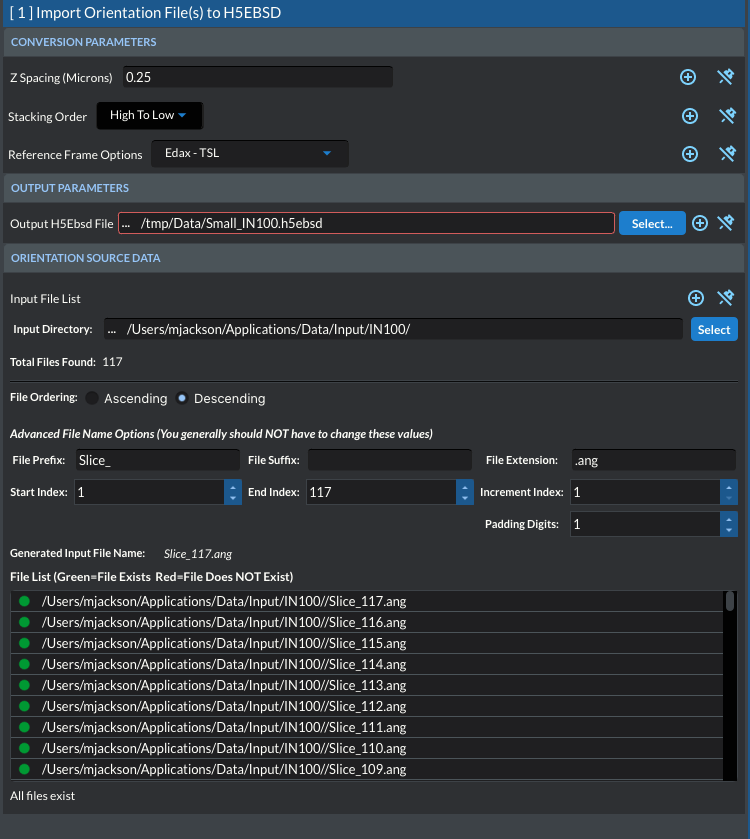
11.32. Import Orientation File(s) to H5EBSD
Group (Subgroup)
IO (Input)
Description
This Filter will convert orientation data obtained from Electron Backscatter Diffraction (EBSD) experiments into a single file archive based on the HDF5 file specification. See the Supported File Formats section below for information on file compatibility. This Filter is typically run as a single Filter Pipeline to perform the conversion. All subsequent Pipelines should then use the Read H5EBSD File Filter to import the H5EBSD file into DREAM3D-NX for analysis, as opposed to re-importing the raw EBSD files. The primary purpose of this Filter is to import a stack of data that forms a 3D volume. If the user wishes to import a single data file, then the Filters Read EDAX EBSD Data (.ang), Read EDAX EBSD Data (.h5), or Read Oxford Instr. EBSD Data (.ctf) should be used for EDAX .ang, EDAX .h5, or Oxford .ctf files, respectively.
Converting Orientation Data to H5EBSD Archive
In order to work with orientation data, DREAM3D-NX needs to read the data from an archive file based on the HDF5 specification. In order to convert the data, the user will first build a single Filter Pipeline by selecting the Import Orientation File(s) to H5EBSD Filter. This Filter will convert a directory of sequentially numbered files into a single HDF5 file that retains all the meta data from the header(s) of the files. The user selects the directory that contains all the files to be imported then uses the additional input widgets on the Filter interface (File Prefix, File Suffix, File Extension, and Padding Digits) to make adjustments to the generated file name until the correct number of files is found. The user may also select starting and ending indices to import. The user interface indicates through red and green icons if an expected file exists on the file system and will also display a warning message at the bottom of the Filter interface if any of the generated file names do not appear on the file system.
Stacking Order
Due to different experimental setups, the definition of the bottom slice or the Z=0 slice can be different. The user should verify that the proper button box is checked for their data set.
Low to High This means that the file with the lowest index, closest to zero (0), will be considered the Z=0 slice.
High to Low This means that the file with the highest index, farthest from zero (0), will be considered the Z=0 slice.
Z Resolution
Many serial sectioning systems are inherently a series of 2D scans stacked together to form a 3D volume of material. Therefore, the experimental systems have no knowledge of the amount of material that was removed between each slice and so the user is responsible for setting this value correctly for their data set.
Orientations, Reference Frames and Coordinate Systems
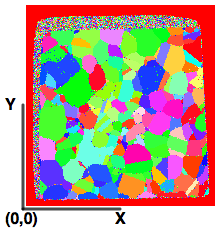
DREAM3D-NX’s origin follows the specimen’s coordinate system so that the physical location of the 0 row and 0 column voxel should visually appear in the lower left corner of a computer graphics display as shown in the figure below, where the specimen coordinate system (white) overlaid with EBSD coordinate system (yellow).

Commercial EBSD acquisition systems do not typically follow this convention, and DREAM3D-NX needs input from the user so that the proper transformations to the data can be applied during subsequent analysis. Commercial EBSD software packages allow for some initial transformations of the data, in which case the DREAM3D-NX environment does not have any way of determining if those transformations have already occurred. During the import process the user is asked a few questions regarding the orientation of the EBSD data in relation to the specimen coordinate system.
Setting the Spatial Reference Frame
The user needs to click the Set Reference Frame button to set the proper reference frame for the data set which will be written to the H5EBSD file as meta data. Below are a number of examples showing the differences in the data sets due to the selection of different reference frames.
No Sample Transform |
Edax/HKL Sample Transform |
|---|---|
|
|
Preset Transformations
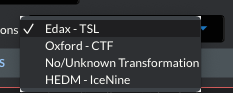
Manufacturer |
Sample Reference Transformation |
Euler Transformation |
|---|---|---|
Edax - TSL |
180 @ <010> |
90 @ <001> |
Oxford - HKL |
180 @ <010> |
0 @ <001> |
No Transform |
0 @ <001> |
0 @ <001> |
HEDM - IceNine |
0 @ <001> |
0 @ <001> |
Supported File Formats
Manufacturer |
File Extension |
Comments |
|---|---|---|
EDAX |
.ang |
– |
Oxford Instruments |
.ctf |
Euler Angles can be stored in Degrees or Radians. |
HEDM from APS |
.mic |
A .config file with the same name is needed for each .mic file |
Completing the Conversion
Once all the inputs are correct the user can click the Go button to start the conversion. Progress will be displayed at the bottom of the DREAM3D user interface during the conversion.
Input Parameter(s)
Parameter Name |
Parameter Type |
Parameter Notes |
Description |
|---|---|---|---|
Z Spacing (Microns) |
Scalar Value |
Float32 |
The spacing between each slice of data |
Stacking Order |
Choices |
The order the files should be placed into the |
|
Reference Frame Options |
Choices |
The reference frame transformation. 0=EDAX(.ang), 1=Oxford(.ctf), 2=No/Unknown Transformation, 3=HEDM-IceNine |
Output File
Parameter Name |
Parameter Type |
Parameter Notes |
Description |
|---|---|---|---|
Output H5Ebsd File |
FileSystemPath |
The path to the generated .h5ebsd file |
Input Data Files
Parameter Name |
Parameter Type |
Parameter Notes |
Description |
|---|---|---|---|
Input File List |
GeneratedFileList |
The values that are used to generate the input file list. See GeneratedFileListParameter for more information. |
Example Pipelines
(01) SmallIN100 Archive
01_Import Data
License & Copyright
Please see the description file distributed with this Plugin
DREAM3D-NX Help
If you need help, need to file a bug report or want to request a new feature, please head over to the DREAM3DNX-Issues GitHub site where the community of DREAM3D-NX users can help answer your questions.